Research
Autophagy, the cellular waste disposal system
Our group aims at understanding how the molecular machinery of autophagy, the cellular waste disposal system, works.
One of the major cellular responses when nutrients are scarce is the activation of a degradation pathway termed autophagy, in which the cell digests its own components. This mechanism provides the cell with nutrients to maintain vital cellular functions during times of starvation. It also serves a house-keeping function by eliminating superfluous or damaged organelles, misfolded proteins, and invading microorganisms, thereby acting as the cellular waste disposal and recycling system. Even though autophagy has been extensively studied from yeast to mammals, the molecular events of starvation sensing and autophagy induction remain elusive. One of the key regulators of autophagy is the target of rapamycin (TOR) kinase, which shuts off autophagy in the presence of nutrients and growth factors. Nutrient-limiting conditions lead to TOR kinase inactivation and autophagy induction, which results in the sequestration of cytosol and organelles into a double-membrane organelle followed by their subsequent delivery to the vacuole/lysosome for breakdown and recycling (Figure 1-3).
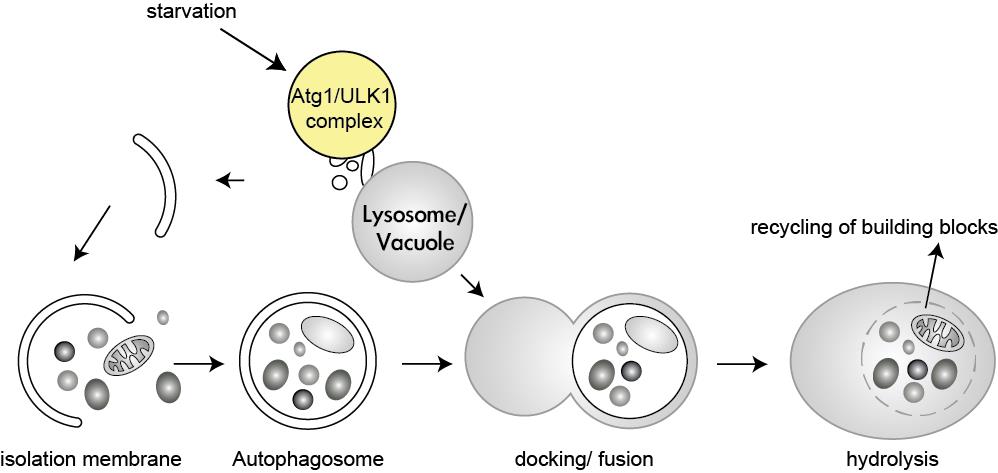
Figure 1: The Atg1/ULK1 kinase complex receives the autophagy-inducing signal, resulting in the initiation of autophagy. An isolation membrane expands and engulfs cytoplasmic material to form a double-membraned autophagosome. After fusion with the lysosome/vacuole the contents of the autophagosome are degraded and recycled for further use.
A major conserved upstream regulator of autophagy is the Atg1 (yeast) / ULK1 (mammals) kinase complex, which is thought to be a direct target of nutrient signaling by TOR. The activity of the Atg1/ULK1 complex is absolutely essential for autophagy function, yet how Atg1/ULK1 regulates autophagosome formation at different stages during the process and the underlying signaling and mechanistic events remain unclear.
We want to dissect the mechanisms by which Atg1/ULK1 kinase activity is regulated and how Atg1/ULK1 itself regulates downstream events in autophagy. We employ a diverse range of classical in vitro and in vivo methods. Recently we have established a complementary synthetic in vivo approach (Torggler et al., Mol Cell 2016). This approach allows us to achieve higher spatial and temporal resolution when studying autophagy in living cells and to define what is necessary and sufficient in vivo at specific steps in autophagy.
Most of our in vivo work is performed in budding yeast, which is an excellent model organism for studying the mechanisms of autophagy due to the highly-conserved nature of the process up to mammals. Additionally, we also use mammalian cell culture systems to test our findings for conservation in higher eukaryotes.
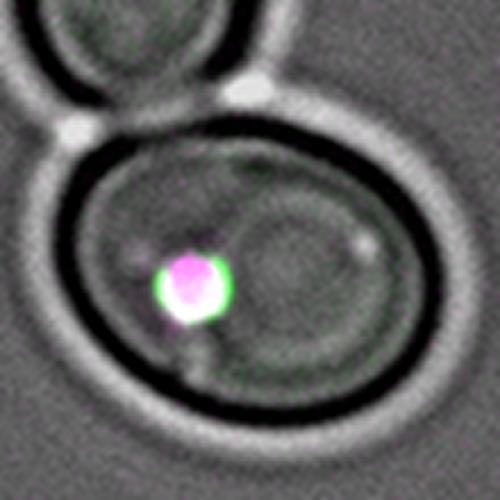 | Figure 2: An over-sized autophagic cargo (magenta) is engulfed by an isolation membrane (green) in a yeast cell, monitored by live cell fluorescence imaging. |
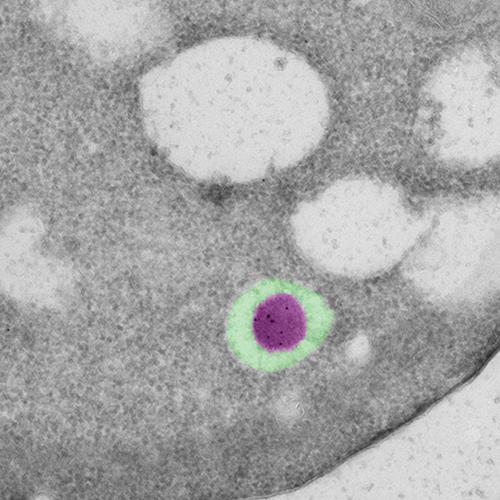 | Figure 3: Native cargo (magenta) in an autophagosome (green) visualized by immuno-electron microscopy. |
Deciphering the mechanisms governing autophagy will help to better understand the molecular basis of diseases associated with autophagy dysfunction, including cancer and neurodegenerative disorders like Alzheimer’s disease.

